Extend ranges for prime editing
extend_for_pe.RdExtend target ranges to span in which to look for spacer-pam seqs
extend_for_pe( gr, bsgenome, nrt = 16, spacer = strrep("N", 20), pam = "NGG", plot = FALSE )
Arguments
| gr | |
|---|---|
| bsgenome | |
| nrt | number: reverse transcription length |
| spacer | string: spacer pattern in extended IUPAC alphabet |
| pam | string: pam pattern in extended IUPAC alphabet |
| plot | TRUE (default) or FALSE |
Value
Details
Extend target ranges to find nearby spacers for prime editing
Examples
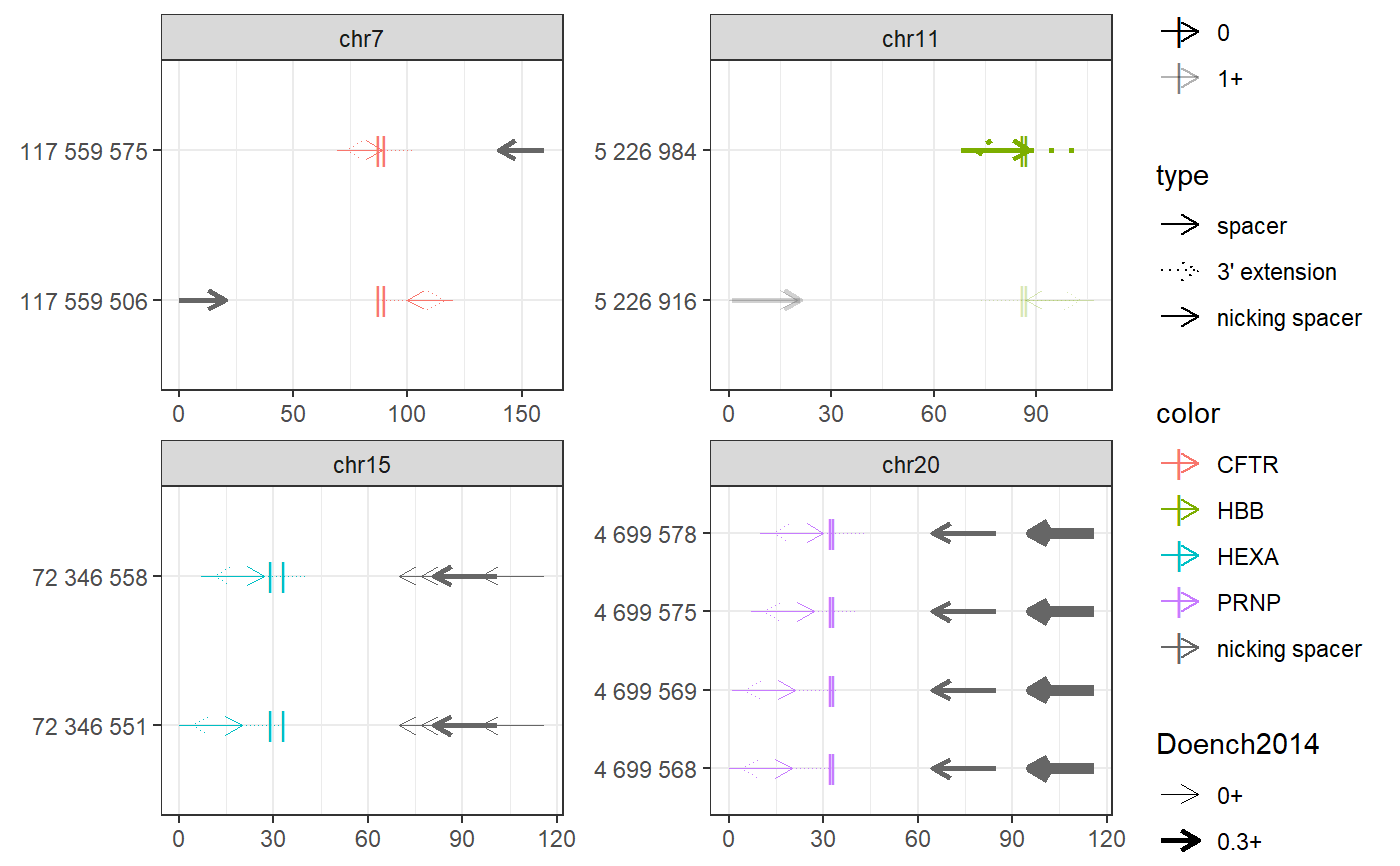
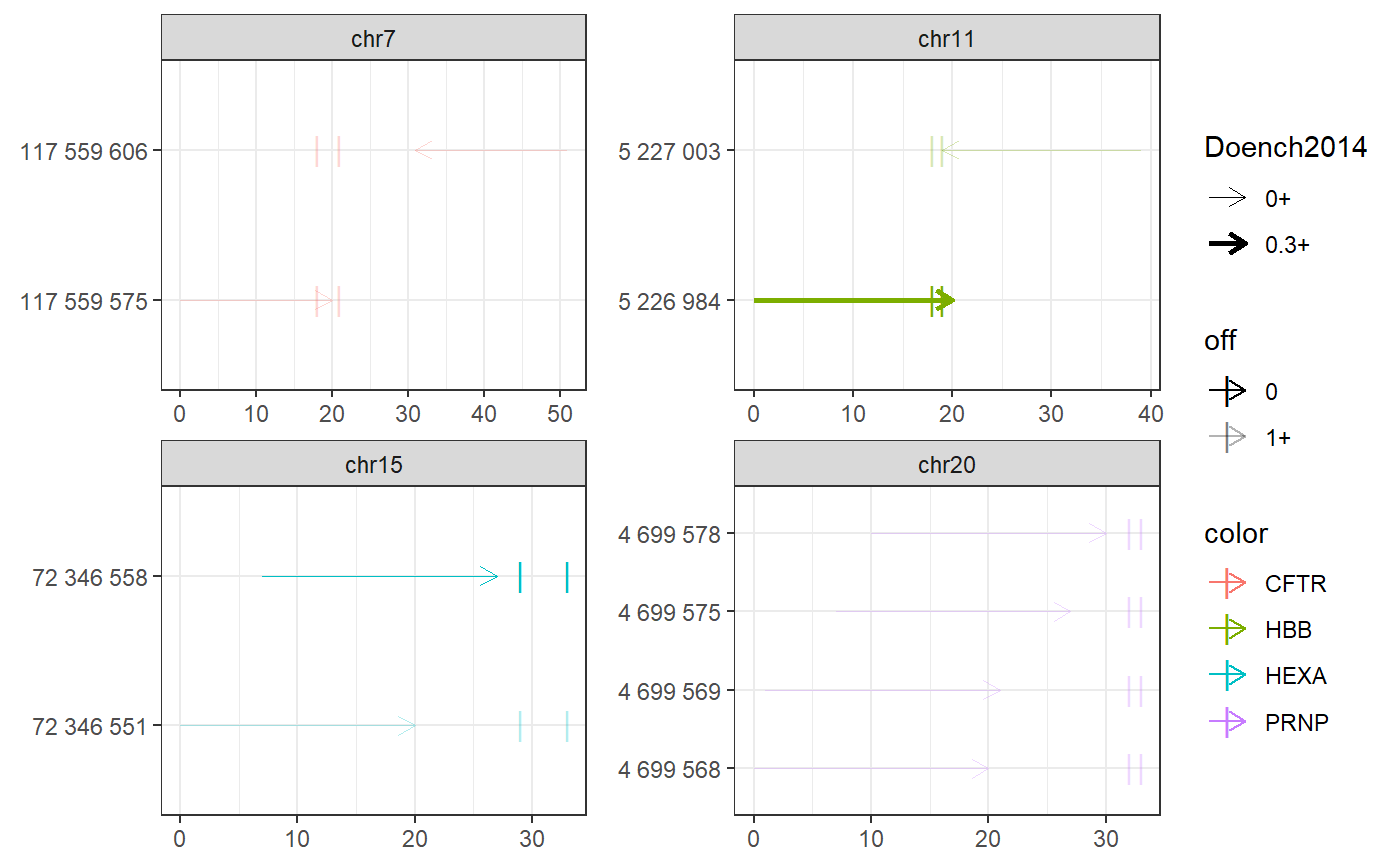
require(magrittr) bsgenome <- BSgenome.Hsapiens.UCSC.hg38::BSgenome.Hsapiens.UCSC.hg38 gr <- char_to_granges(c( PRNP = 'chr20:4699600:+', # snp HBB = 'chr11:5227002:-', # snp HEXA = 'chr15:72346580-72346583:-', # del CFTR = 'chr7:117559593-117559595:+'), # ins bsgenome = bsgenome) find_primespacers(gr, bsgenome)#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#> Warning: Removed 34 rows containing missing values (geom_point).#> Warning: Removed 34 rows containing missing values (geom_point).#> GRanges object with 10 ranges and 21 metadata columns: #> seqnames ranges strand | targetname targetstart #> <Rle> <IRanges> <Rle> | <character> <integer> #> CFTR_1 chr7 117559575-117559594 + | CFTR 117559593 #> CFTR_2 chr7 117559606-117559625 - | CFTR 117559593 #> HBB_1 chr11 5227003-5227022 - | HBB 5227002 #> HBB_2 chr11 5226984-5227003 + | HBB 5227002 #> HEXA_1 chr15 72346551-72346570 + | HEXA 72346580 #> HEXA_2 chr15 72346558-72346577 + | HEXA 72346580 #> PRNP_1 chr20 4699568-4699587 + | PRNP 4699600 #> PRNP_2 chr20 4699569-4699588 + | PRNP 4699600 #> PRNP_3 chr20 4699575-4699594 + | PRNP 4699600 #> PRNP_4 chr20 4699578-4699597 + | PRNP 4699600 #> targetend crisprname crisprspacer crisprpam crisprprimer #> <integer> <character> <character> <character> <character> #> CFTR_1 117559595 CFTR_1 ATTAAAGAAAATATCATCTT TGG AAGAAAATATCAT #> CFTR_2 117559595 CFTR_2 TCTGTATCTATATTCATCAT AGG TATCTATATTCAT #> HBB_1 5227002 HBB_1 CATGGTGCATCTGACTCCTG AGG GTGCATCTGACTC #> HBB_2 5227002 HBB_2 GTAACGGCAGACTTCTCCTC AGG CGGCAGACTTCTC #> HEXA_1 72346583 HEXA_1 TGTAGAAATCCTTCCAGTCA GGG GAAATCCTTCCAG #> HEXA_2 72346583 HEXA_2 ATCCTTCCAGTCAGGGCCAT AGG TTCCAGTCAGGGC #> PRNP_1 4699600 PRNP_1 AGCAGCTGGGGCAGTGGTGG GGG GCTGGGGCAGTGG #> PRNP_2 4699600 PRNP_2 GCAGCTGGGGCAGTGGTGGG GGG CTGGGGCAGTGGT #> PRNP_3 4699600 PRNP_3 GGGGCAGTGGTGGGGGGCCT TGG CAGTGGTGGGGGG #> PRNP_4 4699600 PRNP_4 GCAGTGGTGGGGGGCCTTGG CGG TGGTGGGGGGCCT #> crisprtranscript crisprextension #> <character> <character> #> CFTR_1 CTTTGGTGTTTCCTAT ATAGGAAACACCAAAGATGATATTTTCTT #> CFTR_2 CATAGGAAACACCAAA TTTGGTGTTTCCTATGATGAATATAGATA #> HBB_1 CTGAGGAGAAGTCTGC GCAGACTTCTCCTCAGGAGTCAGATGCAC #> HBB_2 CTCAGGAGTCAGATGC GCATCTGACTCCTGAGGAGAAGTCTGCCG #> HEXA_1 TCAGGGCCATAGGATA TATCCTATGGCCCTGACTGGAAGGATTTC #> HEXA_2 CATAGGATATACGGTT AACCGTATATCCTATGGCCCTGACTGGAA #> PRNP_1 TGGGGGGCCTTGGCGG CCGCCAAGGCCCCCCACCACTGCCCCAGC #> PRNP_2 GGGGGGCCTTGGCGGC GCCGCCAAGGCCCCCCACCACTGCCCCAG #> PRNP_3 CCTTGGCGGCTACATG CATGTAGCCGCCAAGGCCCCCCACCACTG #> PRNP_4 TGGCGGCTACATGCTG CAGCATGTAGCCGCCAAGGCCCCCCACCA #> crisprextrange off off0 Doench2014 #> <character> <numeric> <numeric> <numeric> #> CFTR_1 chr7:117559579-117559607:- 0 0 0.03399990 #> CFTR_2 chr7:117559593-117559621:+ 0 0 0.22337056 #> HBB_1 chr11:5226990-5227018:+ 1 1 0.13111422 #> HBB_2 chr11:5226988-5227016:- 0 0 0.41398065 #> HEXA_1 chr15:72346555-72346583:- 0 0 0.10728361 #> HEXA_2 chr15:72346562-72346590:- 0 0 0.03942528 #> PRNP_1 chr20:4699572-4699600:- 0 0 0.00994899 #> PRNP_2 chr20:4699573-4699601:- 0 0 0.04496627 #> PRNP_3 chr20:4699579-4699607:- 0 0 0.00996254 #> PRNP_4 chr20:4699582-4699610:- 0 0 0.20935863 #> nickrange #> <character> #> CFTR_1 chr7:117559646-117559665:- #> CFTR_2 chr7:117559506-117559525:+ #> HBB_1 chr11:5226916-5226935:+;chr11:5226917-5226936:+ #> HBB_2 <NA> #> HEXA_1 chr15:72346621-72346640:-;chr15:72346628-72346647:-;chr15:72346632-72346651:-;chr15:72346647-72346666:- #> HEXA_2 chr15:72346621-72346640:-;chr15:72346628-72346647:-;chr15:72346632-72346651:-;chr15:72346647-72346666:- #> PRNP_1 chr20:4699632-4699651:-;chr20:4699633-4699652:-;chr20:4699664-4699683:- #> PRNP_2 chr20:4699632-4699651:-;chr20:4699633-4699652:-;chr20:4699664-4699683:- #> PRNP_3 chr20:4699632-4699651:-;chr20:4699633-4699652:-;chr20:4699664-4699683:- #> PRNP_4 chr20:4699632-4699651:-;chr20:4699633-4699652:-;chr20:4699664-4699683:- #> nickspacer #> <character> #> CFTR_1 AGTTTCTTACCTCTTCTAGT #> CFTR_2 GAGGGTAAAATTAAGCACAG #> HBB_1 ACCTTGATACCAACCTGCCC;CCTTGATACCAACCTGCCCA #> HBB_2 <NA> #> HEXA_1 TGGTCACCAAGGCCGGCTTC;CTGGAACTGGTCACCAAGGC;GGAGCTGGAACTGGTCACCA;AGTGAACTATATGAAGGAGC #> HEXA_2 TGGTCACCAAGGCCGGCTTC;CTGGAACTGGTCACCAAGGC;GGAGCTGGAACTGGTCACCA;AGTGAACTATATGAAGGAGC #> PRNP_1 TCACTGCCGAAATGTATGAT;GTCACTGCCGAAATGTATGA;GCATGTTTTCACGATAGTAA #> PRNP_2 TCACTGCCGAAATGTATGAT;GTCACTGCCGAAATGTATGA;GCATGTTTTCACGATAGTAA #> PRNP_3 TCACTGCCGAAATGTATGAT;GTCACTGCCGAAATGTATGA;GCATGTTTTCACGATAGTAA #> PRNP_4 TCACTGCCGAAATGTATGAT;GTCACTGCCGAAATGTATGA;GCATGTTTTCACGATAGTAA #> nickpam nickoff nickoff0 nickoff1 nickoff2 #> <character> <character> <character> <character> <character> #> CFTR_1 TGG 0 0 0 0 #> CFTR_2 TGG 0 0 0 0 #> HBB_1 AGG;GGG 1;1 1;1 0;0 0;0 #> HBB_2 <NA> <NA> <NA> <NA> <NA> #> HEXA_1 CGG;CGG;AGG;TGG 0;0;0;0 0;0;0;0 0;0;0;0 0;0;0;0 #> HEXA_2 CGG;CGG;AGG;TGG 0;0;0;0 0;0;0;0 0;0;0;0 0;0;0;0 #> PRNP_1 GGG;TGG;CGG 0;0;0 0;0;0 0;0;0 0;0;0 #> PRNP_2 GGG;TGG;CGG 0;0;0 0;0;0 0;0;0 0;0;0 #> PRNP_3 GGG;TGG;CGG 0;0;0 0;0;0 0;0;0 0;0;0 #> PRNP_4 GGG;TGG;CGG 0;0;0 0;0;0 0;0;0 0;0;0 #> nickDoench2014 #> <character> #> CFTR_1 0.38 #> CFTR_2 0.49 #> HBB_1 0.06;0.42 #> HBB_2 <NA> #> HEXA_1 0.01;0.05;0.32;0.2 #> HEXA_2 0.01;0.05;0.32;0.2 #> PRNP_1 0.14;0.32;0.52 #> PRNP_2 0.14;0.32;0.52 #> PRNP_3 0.14;0.32;0.52 #> PRNP_4 0.14;0.32;0.52 #> ------- #> seqinfo: 595 sequences (1 circular) from hg38 genome(grext <- extend_for_pe(gr))#> GRanges object with 8 ranges and 3 metadata columns: #> seqnames ranges strand | targetname targetstart #> <Rle> <IRanges> <Rle> | <character> <integer> #> PRNP_f chr20 4699568-4699605 + | PRNP 4699600 #> HBB_f chr11 5226970-5227007 + | HBB 5227002 #> HEXA_f chr15 72346551-72346585 + | HEXA 72346580 #> CFTR_f chr7 117559563-117559598 + | CFTR 117559593 #> PRNP_r chr20 4699595-4699632 - | PRNP 4699600 #> HBB_r chr11 5226997-5227034 - | HBB 5227002 #> HEXA_r chr15 72346578-72346612 - | HEXA 72346580 #> CFTR_r chr7 117559590-117559625 - | CFTR 117559593 #> targetend #> <integer> #> PRNP_f 4699600 #> HBB_f 5227002 #> HEXA_f 72346583 #> CFTR_f 117559595 #> PRNP_r 4699600 #> HBB_r 5227002 #> HEXA_r 72346583 #> CFTR_r 117559595 #> ------- #> seqinfo: 595 sequences (1 circular) from hg38 genome#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#>#> GRanges object with 10 ranges and 11 metadata columns: #> seqnames ranges strand | targetname targetstart #> <Rle> <IRanges> <Rle> | <character> <integer> #> CFTR_f chr7 117559575-117559594 + | CFTR 117559593 #> CFTR_r chr7 117559606-117559625 - | CFTR 117559593 #> HBB_f chr11 5226984-5227003 + | HBB 5227002 #> HBB_r chr11 5227003-5227022 - | HBB 5227002 #> HEXA_f_1 chr15 72346551-72346570 + | HEXA 72346580 #> HEXA_f_2 chr15 72346558-72346577 + | HEXA 72346580 #> PRNP_f_1 chr20 4699568-4699587 + | PRNP 4699600 #> PRNP_f_2 chr20 4699569-4699588 + | PRNP 4699600 #> PRNP_f_3 chr20 4699575-4699594 + | PRNP 4699600 #> PRNP_f_4 chr20 4699578-4699597 + | PRNP 4699600 #> targetend crisprname crisprspacer crisprpam off #> <integer> <character> <character> <character> <numeric> #> CFTR_f 117559595 CFTR_f ATTAAAGAAAATATCATCTT TGG 2 #> CFTR_r 117559595 CFTR_r TCTGTATCTATATTCATCAT AGG 1 #> HBB_f 5227002 HBB_f GTAACGGCAGACTTCTCCTC AGG 0 #> HBB_r 5227002 HBB_r CATGGTGCATCTGACTCCTG AGG 4 #> HEXA_f_1 72346583 HEXA_f_1 TGTAGAAATCCTTCCAGTCA GGG 1 #> HEXA_f_2 72346583 HEXA_f_2 ATCCTTCCAGTCAGGGCCAT AGG 0 #> PRNP_f_1 4699600 PRNP_f_1 AGCAGCTGGGGCAGTGGTGG GGG 11 #> PRNP_f_2 4699600 PRNP_f_2 GCAGCTGGGGCAGTGGTGGG GGG 27 #> PRNP_f_3 4699600 PRNP_f_3 GGGGCAGTGGTGGGGGGCCT TGG 11 #> PRNP_f_4 4699600 PRNP_f_4 GCAGTGGTGGGGGGCCTTGG CGG 4 #> off0 off1 off2 Doench2014 #> <numeric> <numeric> <numeric> <numeric> #> CFTR_f 0 0 2 0.03399990 #> CFTR_r 0 0 1 0.22337056 #> HBB_f 0 0 0 0.41398065 #> HBB_r 1 0 3 0.13111422 #> HEXA_f_1 0 0 1 0.10728361 #> HEXA_f_2 0 0 0 0.03942528 #> PRNP_f_1 0 0 11 0.00994899 #> PRNP_f_2 0 9 18 0.04496627 #> PRNP_f_3 0 0 11 0.00996254 #> PRNP_f_4 0 0 4 0.20935863 #> ------- #> seqinfo: 595 sequences (1 circular) from hg38 genome