Extract subranges
extract_subranges.RdExtract subranges from a GRanges-class object
extract_subranges(gr, ir, plot = FALSE)
Arguments
| gr | |
|---|---|
| ir |
|
| plot | TRUE or FALSE (default) |
Value
Examples
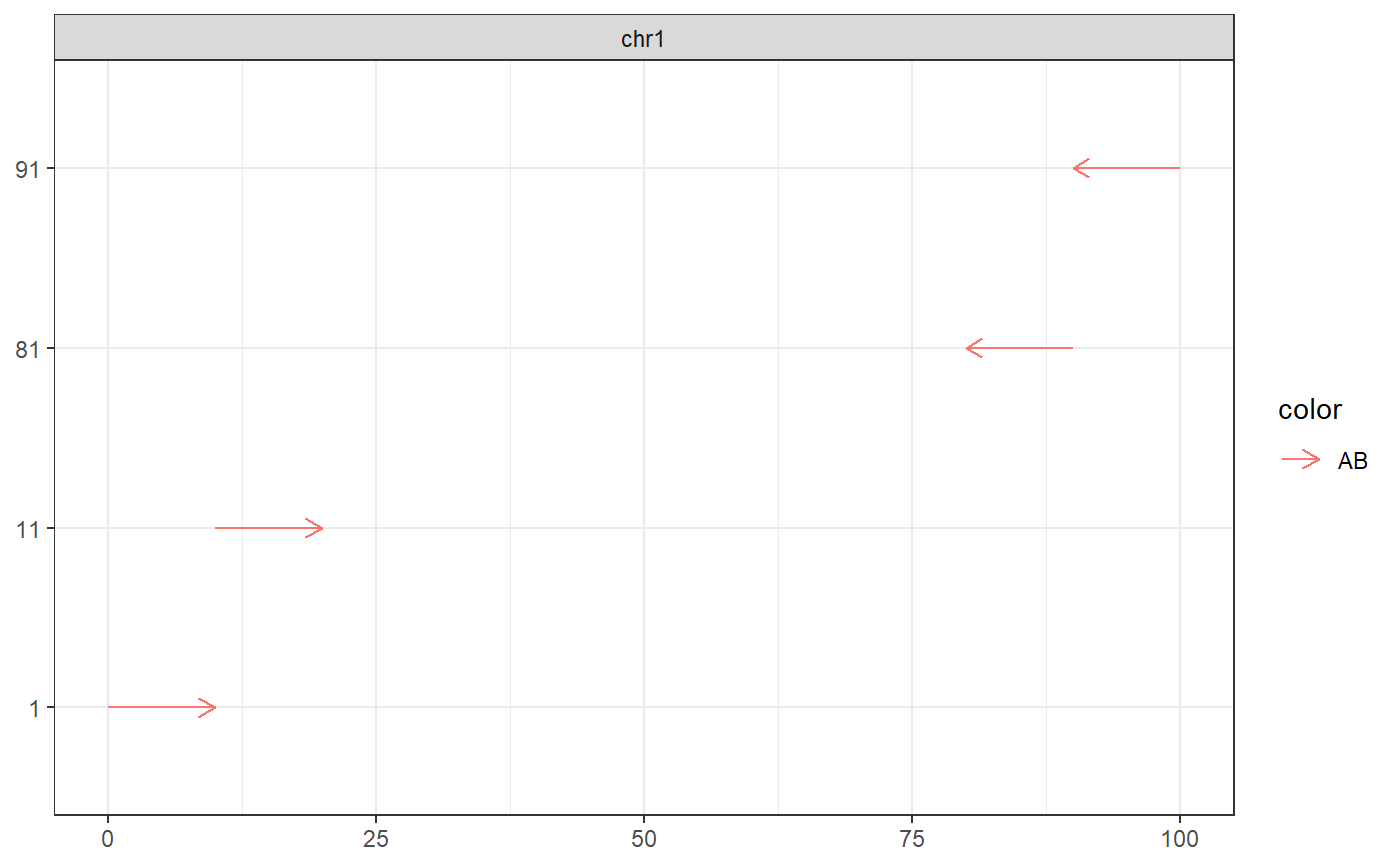
# Extract a subrange gr <- GenomicRanges::GRanges(c(A = 'chr1:1-100:+', B = 'chr1:1-100:-')) gr$targetname <- 'AB' ir <- IRanges::IRanges(c(A = '1-10', A = '11-20', B = '1-10', B = '11-20')) extract_subranges(gr, ir, plot = TRUE)#> GRanges object with 4 ranges and 1 metadata column: #> seqnames ranges strand | targetname #> <Rle> <IRanges> <Rle> | <character> #> A_1 chr1 1-10 + | AB #> A_2 chr1 11-20 + | AB #> B_1 chr1 91-100 - | AB #> B_2 chr1 81-90 - | AB #> ------- #> seqinfo: 1 sequence from an unspecified genome; no seqlengths# Return empty GRanges for empty IRanges extract_subranges(GenomicRanges::GRanges('chr1:345-456'), IRanges::IRanges())#> GRanges object with 0 ranges and 0 metadata columns: #> seqnames ranges strand #> <Rle> <IRanges> <Rle> #> ------- #> seqinfo: 1 sequence from an unspecified genome; no seqlengths